There was a time, not that long ago, when 1-3% ADAR editing rates in tissue culture cells were typically reported in the field. The hope then was that with further chemical optimization, editing rates could be increased high enough to have a clinically relevant impact in the setting of a gain-of-function approach. Mathematically speaking, go from nothing to something is an immeasurable relative increase.
In the realm of biology, this is pertinent to diseases like Duchenne Muscular Dystrophy or Spinal Muscular Atrophy where
relatively small, 10-20% target engagement by RNA Therapeutics have been demonstrated (splice modulator SPINRAZA) or are
expected (exon skippers) to have big disease modifying activity when given to patients essentially genetically null (=not expressing) dystrophin and SMN1, respectively.
In theory,
the upper limit of ADAR Editing should be extremely high since near-complete editing of ion channel and neurotransmitter receptor pre-mRNAs are seen in neurobiology.
The
perception that ADAR Editing mediated by oligonucleotides could be generally
low in clinical applications started to shift when Monian and colleagues at
Wave Life Sciences reported in a groundbreaking Nature Biotech paper a year ago
robust 50%+ editing rates of the alpha-1 antitrypsin Z allele.
It suggested that this can be
achieved by painstaking chemical optimization at a ‘lucky' target site. This is not much different from how small
molecules get chemically matured following an initial low-affinity,
low-specificity hit.
For the
type of blockbuster market opportunity like alpha-1-antitrypsin this effort is well worth it. Still, finding potent editing-enabling oligos in an efficient manner would
open many doors such as testing scientific hypotheses faster, especially as
the ADAR Editing pioneers sift through their list of candidate targets and
indications.
Enter Wave
Life Sciences and their PRISM platform.
I have never quite understood what exactly is behind this platform for
oligonucleotide discovery (RNAi, exon
skipping and ADAR Editing). It does appear,
however, to test many combinations of chemical modifications at the
base, sugar, and internucleotide linker while considering their chemical
neighborhoods and stereochemistry, ultimately coming up with design principles for potent oligonucleotide
therapeutics candidates.
Certainly,
given that modern lab automation allows for increased throughputs (see
biotechtv tour of Aera Therapeutics), such a Big Data, Artificial Intelligence approach to
oligonucleotide drug development makes a lot of sense and can give biotech
companies crucial competitive advantages in terms of time and better molecules.
Two
chemistry insights regarding ADAR technology that have emerged from PRISM stand out so far. Firstly, the zwitterionic phosphorylguanidine
(PN) backbone linker, preferably in a stereopure format. The PN chemistry is shown to allow for improved unassisted cellular uptake into various cell types in mice (immune cells,
various liver cell types, renal cells etc) while stereopurity brings advantages
in terms of ADAR recognition of the duplex substrate. This sounds similar to the morpholino chemistry that has proven to be quite safe following systemic administration (e.g. eteplirsen by Sarepta), but is very rapidly
eliminated from the circulation into urine. It will therefore be important to show in future
studies that the dose demands for oligos with predominantly PN backbone are not as high.
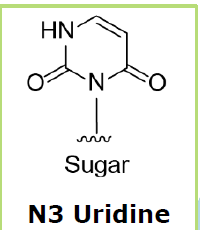
With regard
to the orphan base, the base opposite the adenosine to be edited, Wave is
honing in on deoxy-N3 uridine as its preferred chemistry.
In addition to alpha-1-antitrypsin (now partnered with GSK), robust 50-90% editing is demonstrated for a number of clinically relevant genes such as UGP2 (à epileptic encephalopathy 83), and Nrf2-Keap signaling (stress regulation in chronic disease). For the latter two targets, not just one, but several highly potent editing oligos could be identified. This reflects increased emphasis by Wave in applying RNA Editing to targets that are not for correcting specific mutations, but where the goal is to increase expression of a protein where it could be helpful, but without necessarily changing its inherent function or sequence.
This allows
Wave to scan for potent editing oligos often along the entire target (pre-)mRNA
instead of being limited to just one site! Also, a number of genetic diseases are caused by a various mutations dispersed throughout a gene so that a mutation-correction approach may require the development of
multiple editing oligos and some sites will not be amenable to AàI approaches at all. In the end, this increases the market potential
of a given oligonucleotide.
In summary,
being able to consistently achieve 50%+ editing rates will be sufficient for
most therapeutic editing approaches.
Going from say 2% of something to 50% is a 25-fold increase, but maxing
out at 100% for another 2x may not give that much of additive benefit. Of course, biological pathways can sometimes
be complex and the responses may not be as linear. At 80%+ editing, diseases caused by dominant-negative
mutations would also come within the realm of therapeutic possibilities of ADAR
editing and this includes liver-related diseases of piZZ alpha-1-antitrypsin,
but here ADAR Editing may face inherently more potent knockdown mechanisms such
as RNAi.
Disclosure: I am long WVE, as I am impressed by the speed with which they have chemically matured editing oligos and their dystrophin exon skipper is showing intriguing early clinical results (RNA, not protein level). However, I have not taken a full position yet as I want to see the company first demonstrate robust target engagement in the clinic (including for the dystrophin exon skipper). So far, all the clinical results have greatly disappointed with claims of 10-20%-type target knockdowns